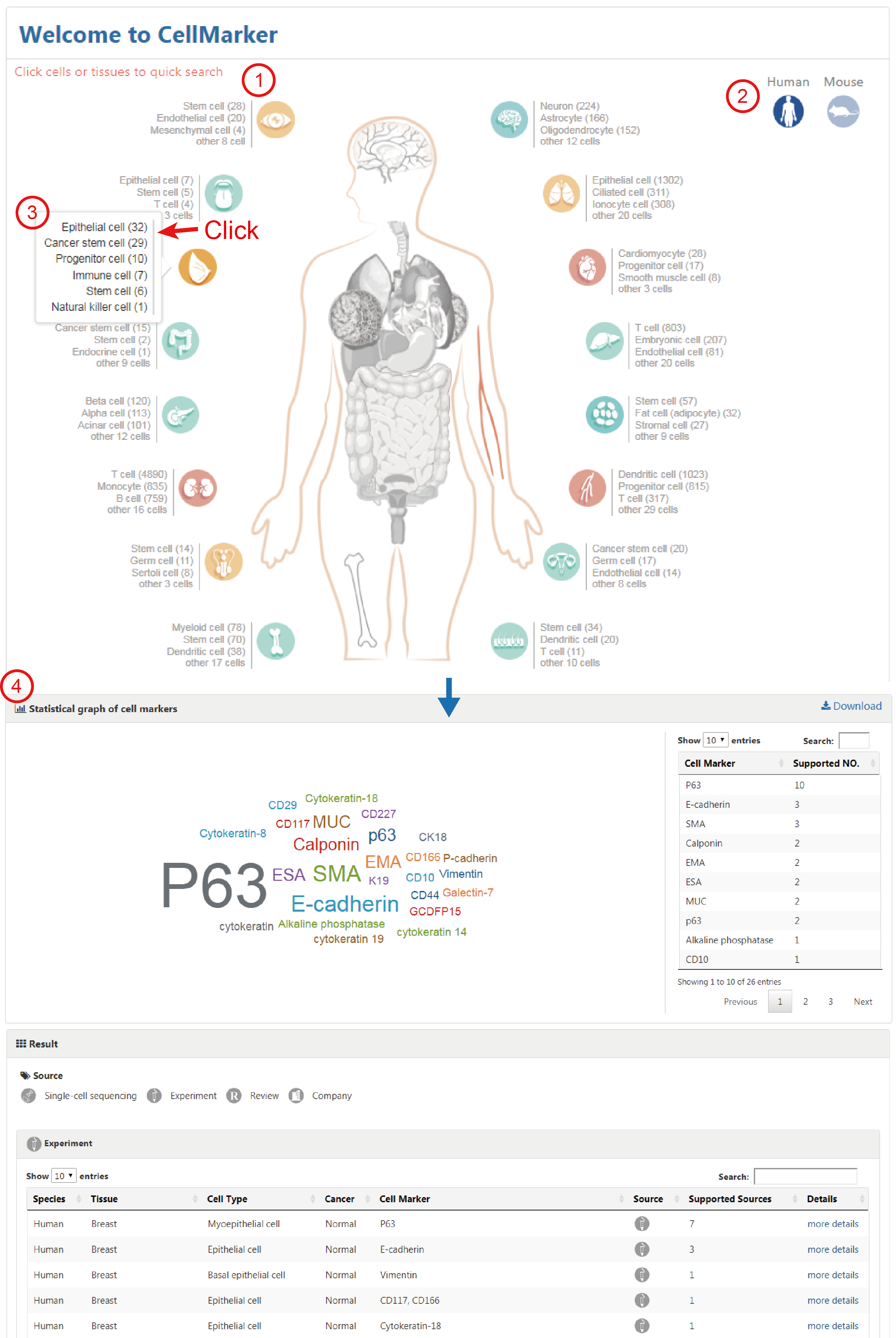
The home page provides a global view of body maps of human and mouse to quickly explore cell markers by clicking on hyperlinks embedded in the web images - 'Anatomical location of human or mouse cells' (1). The mouse body map can be switched by clicking the icon at the top right (2). The body maps can facilitate quick browse of cell markers for the listed cell types and more detailed cell types can be shown by clicking the tissue icons (3). When click a cell type, it will jump to the corresponding search result of the cell markers related to the cell type (4).
The right is a quick search box. Users can search cell markers by inputting cell name, cell marker name, or tissue name of interest.
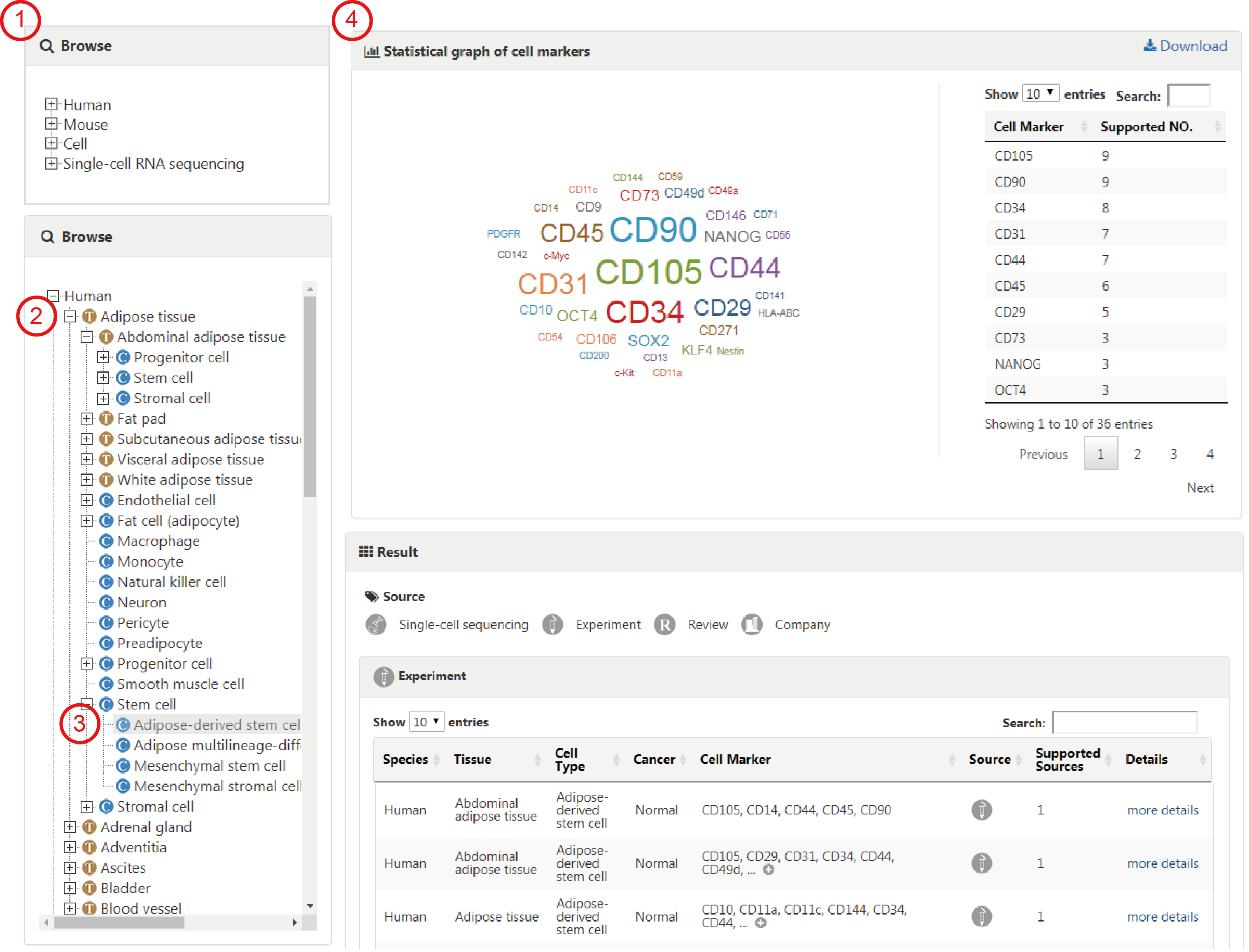
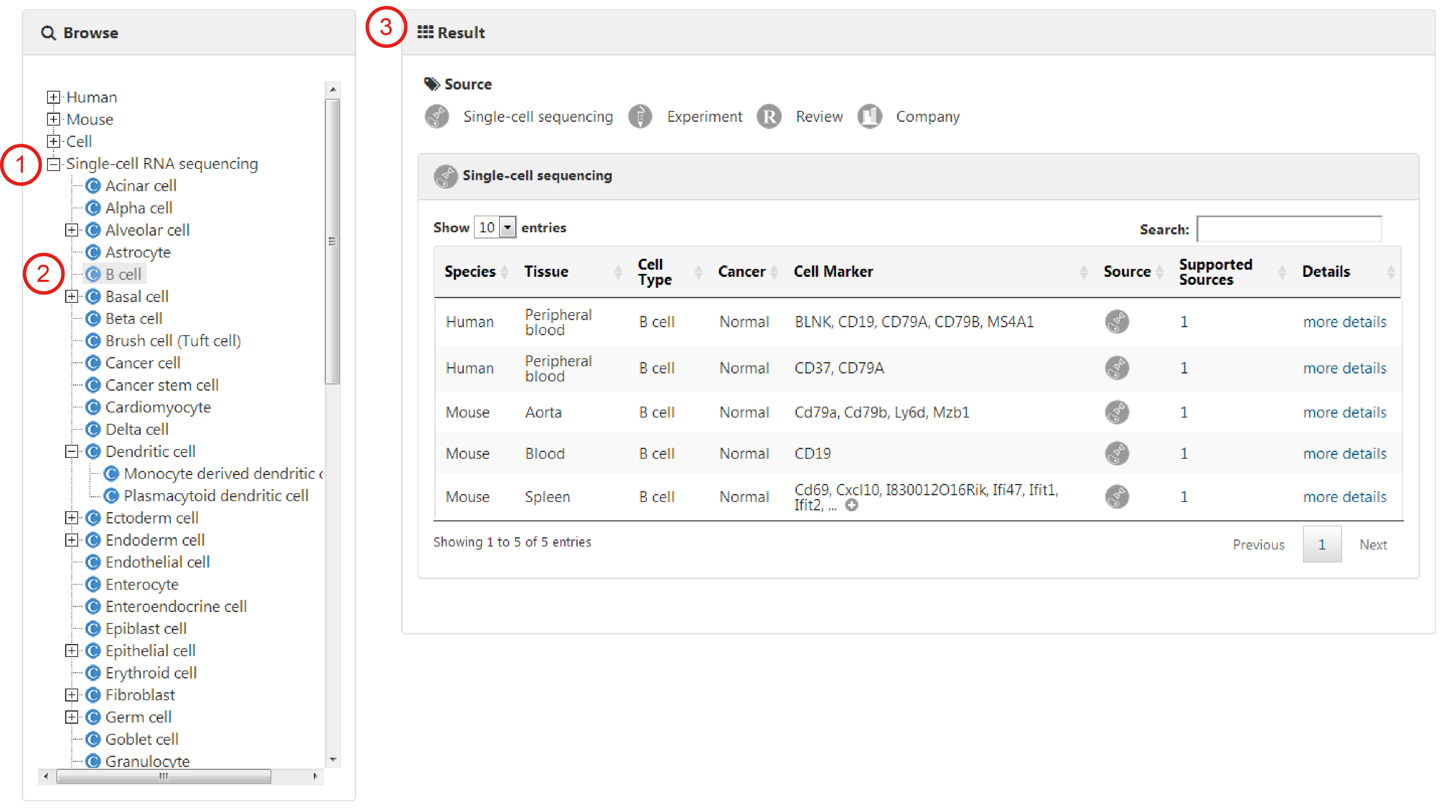
The browse page presents a hierarchical classification of cells and tissues. Users can browse cell markers by clicking cell types in different tissues of human (or mouse), and the complete list of matched cell marker entries can be returned (1-4). For example, to browse the entries related to Adipose tissue in human, you can click 'Human', choose the 'Adipose tissue' (2) and find the interested cell type, such as 'Adipose-derived stem cell' (3). The related cell markers will be shown on the right panel, including a statistical graph of the cell markers for the cell type and the entries from different sources (4).
Users can also browse all entries associated with an interested cell type by directly selecting the corresponding cell (1-3).
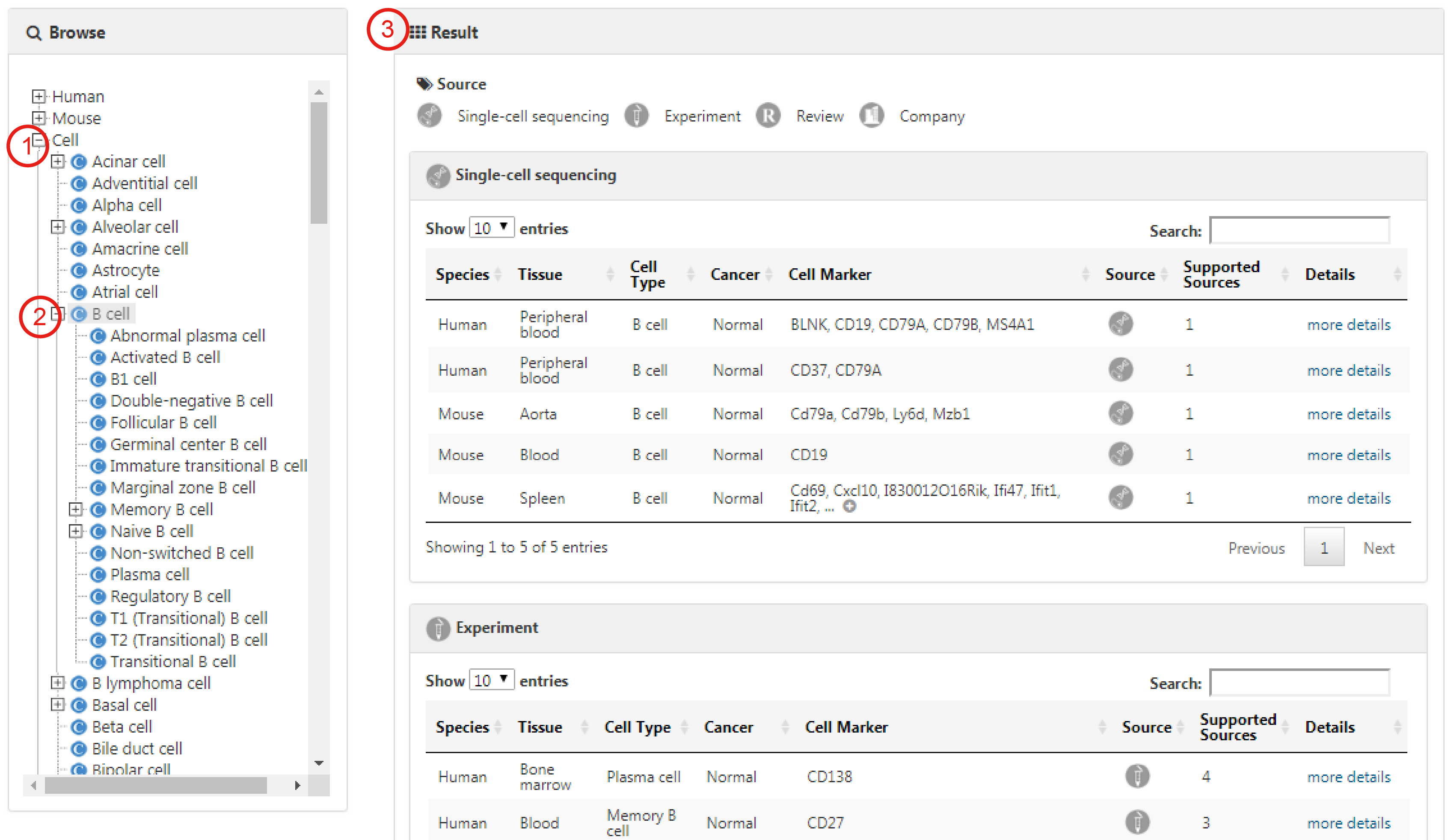
Cell marker entries derived from single-cell sequencing researches are listed separately on the bottom of the browse list.
Users can query the database through two search panels: Cell Search and Marker Search.
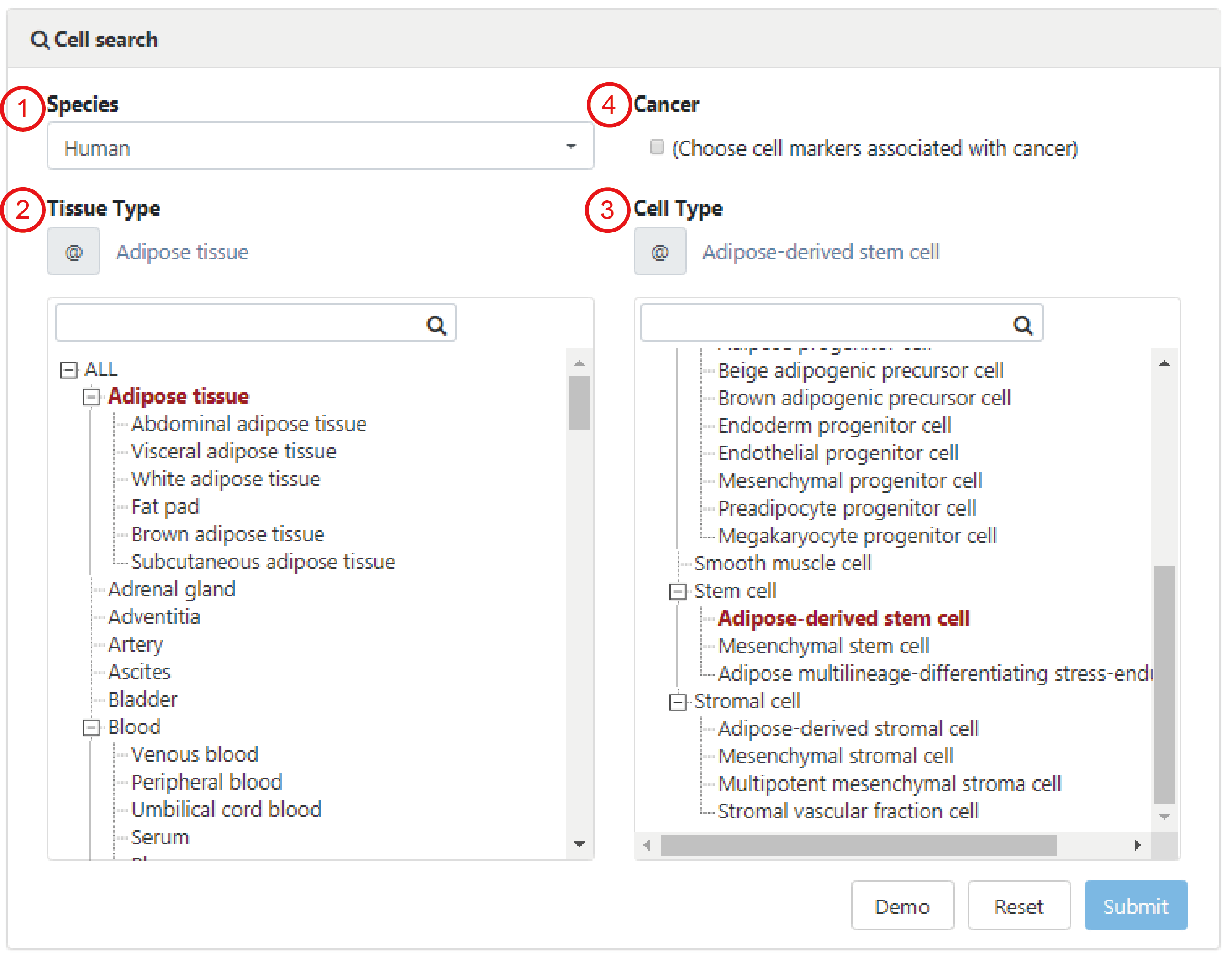
Users can search for cell markers of cells of interest by selecting species from the pull-down menu, tissue type from the hierarchical tree of tissues and cell type from the hierarchical trees of cell types (1-3). When input key words into corresponding searcd box, the candidate tissues or cells stored in the database will be listed for further selection. In particular, cell markers associated with cancers in the corresponding tissue will be returned if choosing the 'Cancer' box (4).
Here, we take 'Adipose-derived stem cell' as an example to explain how to search cell markers in a particular cell. First, choose 'Human' in the 'Species type' search box (1). Next, choose the 'Adipose tissue' in the 'Tissue type' box (2) and then input or choose 'Adipose-derived stem cell' in the 'Cell type' box (3). After clicking on the 'Submit' button, the Result page is displayed.
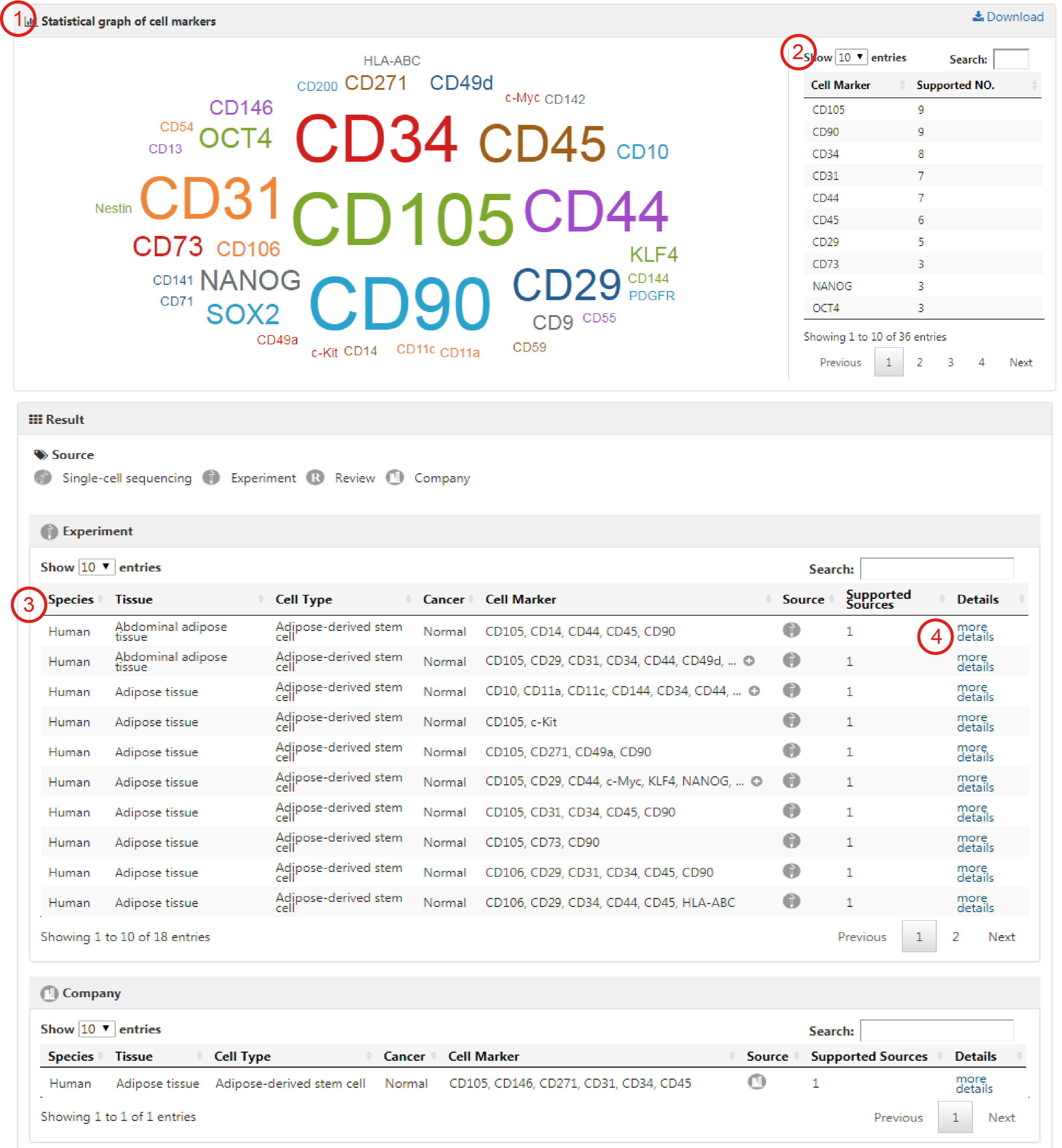
The result page contains an intuitive statistical graph of cell marker prevalence (1), which is quantified by the number of supportive evidence (i.e., publications) and an integrative cell marker list for the queried cell type (2). The detail data can be downloaded by the download icon. Also, the Result page displays four detailed tables of cell marker entries derived from different sources including single-cell RNA sequencing, experiment, review and company. Each entry contains Species, Tissue, Cell Type, Cancer, Cell Marker, Source and Supported Sources (3). By clicking on the hyperlink 'more details' for an individual entry in these tables, users can obtain more detailed information including gene symbols, gene ids, protein names, protein ids, publication information (i.e., title, PubMed ID, journal and publication year) and the cross references to external databases (4). In addition, CellMarker provides several features for browsing the query results. Users can filter entries by typing terms in the 'Search' box at the top right of the table, sort columns in an ascending (or a descending) fashion by clicking the arrows in the column headers and select table size per page by using the 'Show entries' pull-down menu on the top left side above the table.
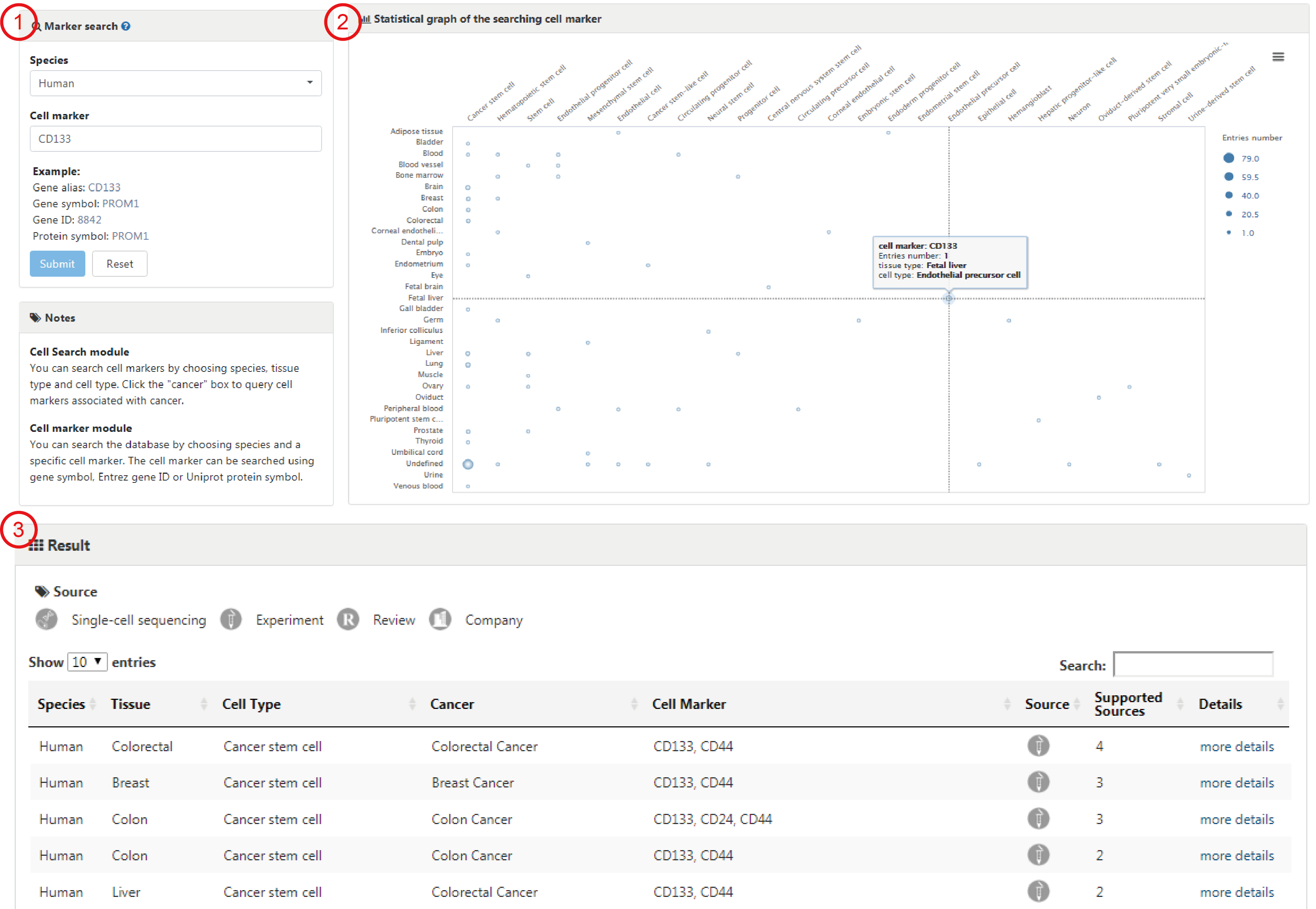
Users can search any genes of interest by entering gene symbols, gene ids or protein names to query in which cell types of which tissues the specific gene can act as cell marker (1). After clicking the 'Submit' button, the search engine will return an interactive bubble chart and a table showing comprehensive information of cell markers (2-3). The interactive bubble chart exhibits how often the gene of interest is used as a cell marker in different cells of different tissues.
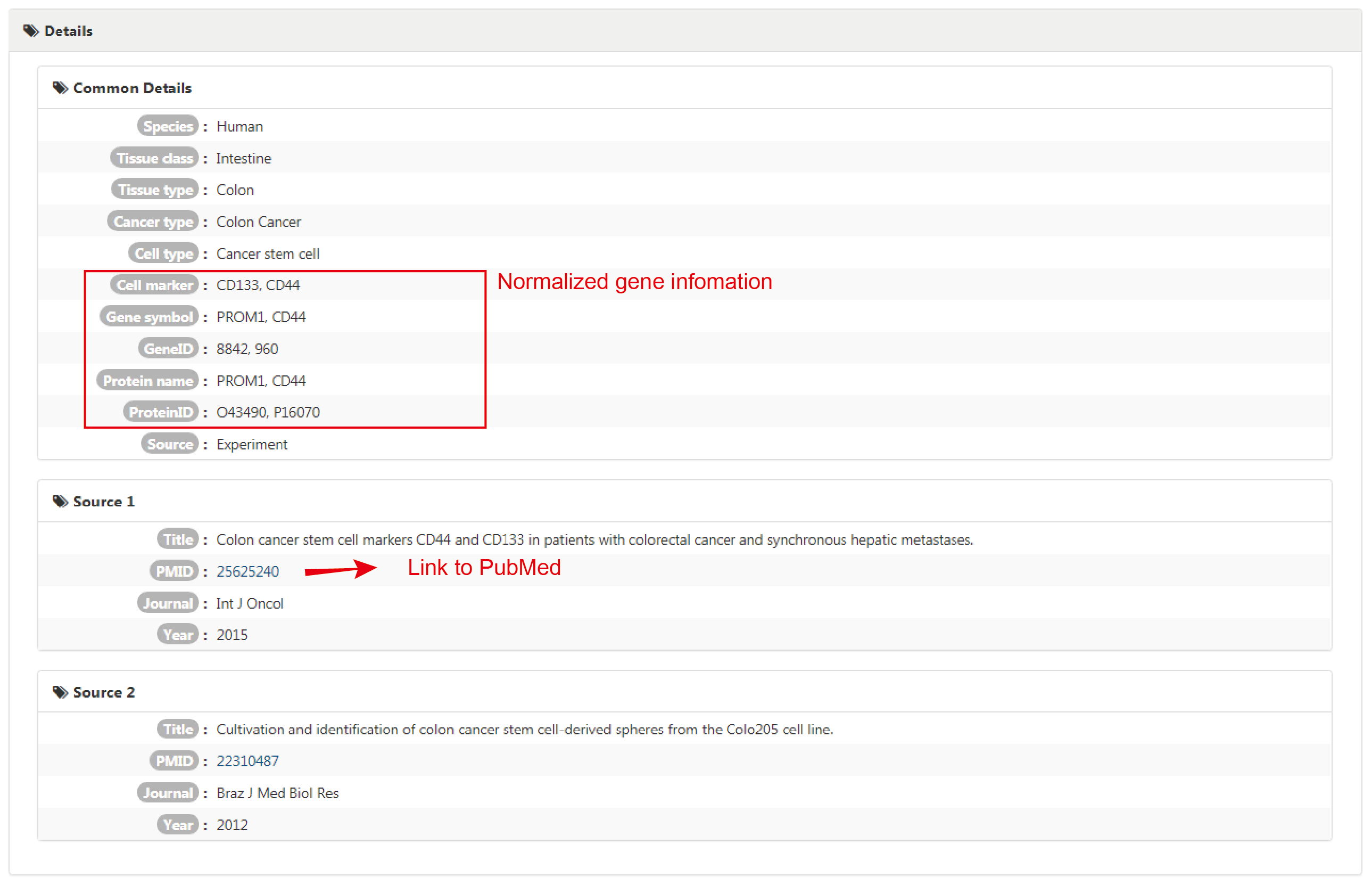
Details interface provides detailed information on each deposited entry of cell markers, which contains comprehensive information of each single record in the entry.
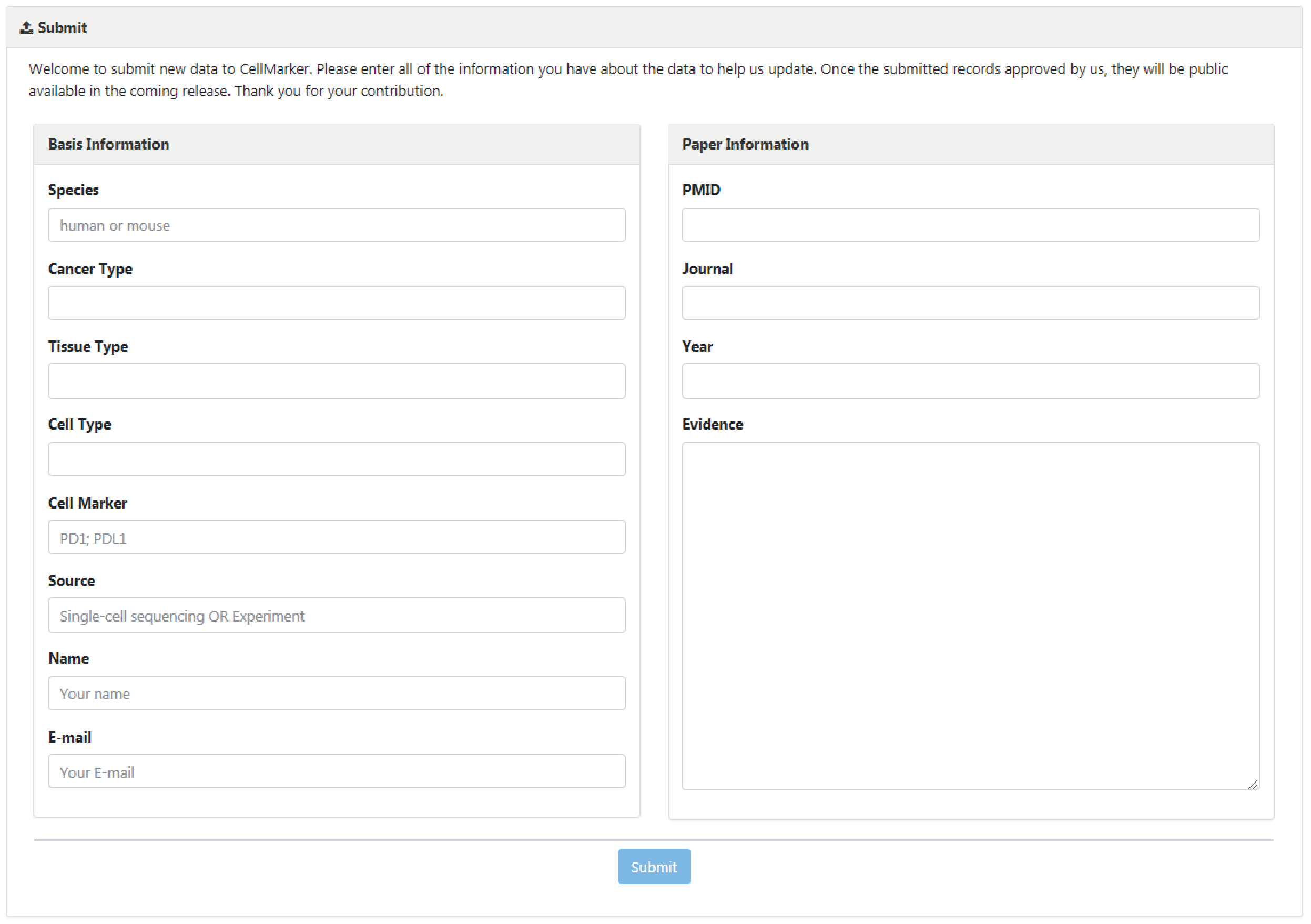
Welcome to submit new data to the CellMarker database. Please enter all of the information you have about the data to help us add a new record. Once the submitted records approved by us, they will be public available in the coming release. Thank you for your contribution.
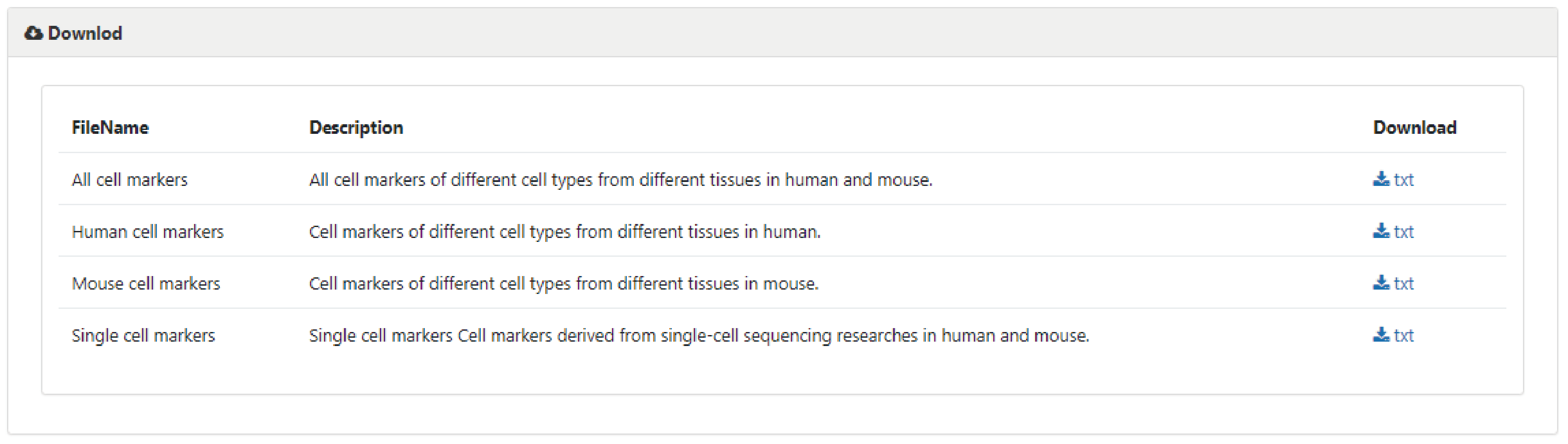
To download data in the database, select the menu 'Download'. CellMarker database provides downloadable file in TXT format. Users can download all cell marker data sets, which include four files: 1) all cell markers of different cell types from different tissues in human and mouse, 2) cell markers in human, 3) cell markers in mouse, and 4) cell markers from single cell sequencing researches.
If you have any questions or comments regarding to CellMarker, you may contact us by sending an email to Yun Xiao (xiaoyun@ems.hrbmu.edu.cn).